Whole-Genome sequencing simulated on supercomputers
(PhysOrg.com) -- The Human Genome Project paved the way for genomics, the study of an organism's genome. Personalized genomics can establish the relationship between DNA sequence variations among individuals and their health conditions and responses to drugs and treatments. To make genome sequencing a routine procedure, however, the time must be reduced to less than a day and the cost to less than $1,000 -- a feat not possible with current knowledge and technologies.
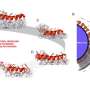
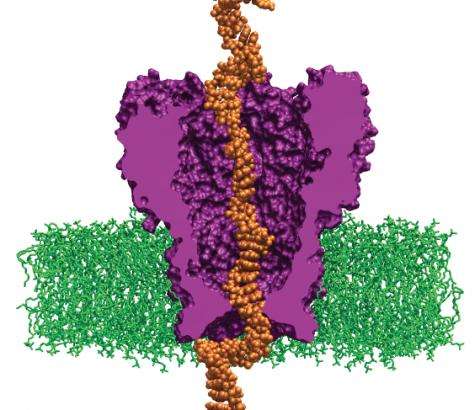
In 2008, a research team led by Aleksei Aksimentiev, assistant professor in the physics department at the University of Illinois-Urbana-Champaign, began a project to create machines for personal genome sequencing that will be more accessible to hospitals. Using Oak Ridge National Laboratory's Jaguar, one of the world's fastest supercomputers, Aksimentiev and his team is developing a nanopore approach, which promises a drastic reduction in time and costs for DNA sequencing. Their research reveals the shape of DNA moving through a single nanopore -- a protein pore a billionth of a meter wide that traverses a membrane. As the DNA passes through the pore, the sequence of nucleotides (DNA building blocks) is read by a detector.
"The main obstacle of sequencing using the older generations of biological and synthetic nanopores was the inability to identify the DNA sequence to single-nucleotide resolution," Aksimentiev said. "The nucleotides passed too quickly through the nanopore for scientists to sequence the DNA."
Aksimentiev's group uses the nanopore MspA, an engineered protein. Its sequence must be altered to bind more strongly to the moving DNA strand. MspA is an ideal platform for sequencing DNA because scientists can now measure dams in the pore, which could slow DNA's journey through the protein. Altering the MspA protein to optimize dams is both time-consuming and costly in a laboratory but simple on a computer. For instance, to alter the protein in any way, scientists must determine whether the particular mutation they introduce is stable and if the idea is reasonable. Therefore, the scientists first simulate MspA to decide on a mutation to induce and to test high-risk ideas before implementing them in an experiment.
The research team uses the code NAMD, which calculates minimum energy states of atoms in a large biomolecular system and is an indicator of what shapes the molecules would be most comfortable assuming. The team first builds a model of the MspA protein submerged in a lipid bilayer and electrolyte solution. A DNA strand of a desired nucleotide sequence is then threaded through the MspA nanopore. Next the scientists simulate the effect of an electric field driving ions and DNA through the MspA nanopore.
The simulation employs molecular dynamics, or calculations of the motion of each atom in a molecular system following the physical laws of nature, to mimic the experimental system. The simulations' results can be directly compared to ones from experiments because both approaches measure the ionic current, according to Aksimentiev. By knowing the positions of each DNA atom and ion, scientists gain an advantage—they can optimize nanopore sequencing using a rational design to produce a pore that hugs to the DNA more tightly, slowing the molecule's journey through the pore to a speed allowing single-nucleotide resolution.
The sequencing work is funded by the National Human Genome Research Institute of the National Institutes of Health. The project's method development is funded in part by the National Science Foundation. Collaborators with the project include two experimental groups: one led by Jens Gundlach at the University of Washington-Seattle and the other by Michael Niederweis at the University of Alabama-Birmingham.
The research received 10 million processor hours on Jaguar through the Innovative and Novel Computational Impact on Theory and Experiment, or INCITE, program, which awards considerable allocations on some of the world's most powerful supercomputers to projects addressing grand challenges in science and engineering. With the INCITE allocation, the scientists were able to reproduce the dams in the MspA nanopore for the type of DNA nucleotides confined to it, slowing down the sequence movement through the nanopore.
"We have carried out a pilot study on several variants of the MspA nanopore and observed considerable reduction of the DNA strand speed," Aksimentiev said. "These very preliminary results suggest that achieving a 100-fold reduction of DNA velocity, which should be sufficient to read out the DNA sequence with single-nucleotide resolution, is within reach. Future studies will be directed toward this goal."
The team hopes to achieve this project's objective by 2013 and plans to pursue a number of exciting spin-off projects, Aksimentiev said. The ability to make genome sequencing affordable will enable such programs as the Cancer Genome Project, which characterizes DNA mutations in cancer cells in various tissues throughout all stages of cancer development.
Provided by Oak Ridge National Laboratory