How microbes take out the trash
(PhysOrg.com) -- The molecular machinery bacteria use to rid themselves of toxic substances including antimicrobial drugs has been studied in detail by a UA-led team of researchers. A better understanding of these mechanisms could lead to new weapons in the fight against pathogens.
Microbes have colonized virtually every spot on this planet, from deep sea vents spewing scalding seawater laden with heavy metals to the icy pinnacles of the world's tallest mountain ranges.
Part of their ability to thrive in the harshest of environments rests on sophisticated cellular mechanisms to deal with substances that are needed to run biochemical processes but dangerous when allowed to accumulate to higher levels.
"Copper is a double-edged sword for organisms because it is a vital ingredient in certain enzymes needed to carry out metabolic functions, but too much of it is toxic and can kill the cell," said Christopher Rensing, an associate professor in the department of soil, water and environmental science in the University of Arizona's College of Agriculture and Life Sciences.
This delicate balance is maintained by specialized transporter proteins embedded in the cell membranes that act as trash men, collecting toxins such as metal ions and pumping them out of the cell.
Research in Rensing's lab focuses on how microbes deal with metals such as copper and silver and discovering new ways to use metals to fight germs.
Scientists have long been fascinated with the intricacy of transporter proteins and their keen ability to recognize chemicals and take the appropriate action. However, many of the precise mechanisms remain elusive.
Studying a class of cellular transporters known as RND proteins in great detail, a team led by UA scientists Eun-Hae Kim, Megan McEvoy and Christopher Rensing has gained new insights into the way they function. Based on the findings, published in the current issue of the Journal of Bacteriology and chosen for a spotlight feature by the journal editors, Kim and her team propose a revised working model for those transporters that has implications for antimicrobial drug development.
"If you're a microbe, the key question is, 'How do you get rid of the excess copper while at the same time keeping enough of it around to sustain your basic life processes?'" Rensing said. "We wanted to find out how the microbes can sense copper levels inside their cells and adjust their pumping action accordingly."
By altering parts of the transporter protein and observing the results, Rensing's doctoral student Eun-Hae Kim discovered that her findings were at odds with the prevailing hypothesis of how the protein transport system worked.
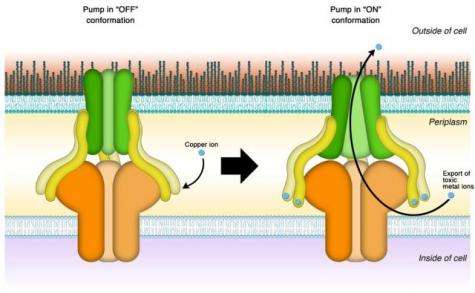
The protein pump consists of several separate protein building blocks that together make up the functional pumping unit. Its two main components are two proteins embedded in each of the two membrane layers enveloping the bacterial cell. A third protein, called the membrane fusion protein, acts as a bridge connecting each of the two membrane proteins. The entire assembly forms a tunnel that, when open, connects the inside of the cell with the outside world.
"People always wondered what the function of that membrane fusion protein was," Kim said. "We knew that in addition to providing the mechanical connection, it actively participates in the pumping process, but nobody had figured out how."
Kim's team hypothesized that rather than binding copper ions one by one and passing them along from one protein component to the next before pushing them out through the tunnel, the membrane fusion protein acts as a power switch that sets a molecular pumping machinery in motion. Accordingly, they called their hypothesis the switch model.
"Once activated, the membrane fusion protein undergoes a change in shape," Kim explained, "This turns on the other components of the transporter protein complex. Previous studies suggest it may open up like an iris before it starts pumping copper ions out of the cell."
In addition to setting the process in motion, Kim's research showed the membrane fusion protein acts as a sensor.
"It has six binding sites for copper," she explained. "Only if enough copper is present to saturate all six does the protein spring into action."
The experimental system developed by Rensing's group serves as a model to study other pump proteins, not just those transporting copper. Similar molecules allow bacteria to eliminate antibiotics and cancer cells to throw out chemotherapeutic drugs.
Understanding the precise molecular workings of pumping proteins in microbes and cells in general has implications spanning global climate, cancer, neurodegenerative diseases such as Alzheimer's and Parkinson's, aging and antibiotics, Rensing said.
"If you know the components, you can develop drugs that target specifically those components and only kill the bacteria that have them, which minimizes side effects."
"Our model allows us to analyze in detail how the pump works, so it becomes possible to develop inhibitors for them," Kim added.
Copper-pumping transporters are also used by macrophages, or white blood cells. They are the immune system's first line of defense against microbes invading the body.
"Macrophages acquire copper ions in an inflamed area to create a chemical milieu favoring the formation of reactive oxygen molecules," Rensing explained. "These reactive oxygen molecules together with the copper ions then kill off the microbes."
Microbes on the other hand have evolved transporter molecules eliminating the toxic copper to help them stave off such attacks.
"If we could block their transporters we would create an Achilles heel rendering them vulnerable to the copper thrown at them by the macrophages."
Before antibiotics became available, many antimicrobial drugs were based on metals, Rensing said.
"Silver in very small amounts inhibits certain enzymes in bacteria," he said. "That's why some of those drugs were called ‘Silver Bullet' or ‘Magic Bullet,'" adding that with more and more microbes becoming resistant to antibiotics, understanding and exploiting molecular pumps as drug targets becomes ever more important.
Rensing's team found RND-type transporter proteins in the membranes of bacteria living in extreme environments such as manganese nodules littering deep sea plains. Even bacteria floating in deep ocean waters, but far above the bottom, rely on metal transporters, as scientists from the Scripps Institution of Oceanography in San Diego, Calif. discovered when they analyzed water samples from the Puerto Rico Trench.
"We were surprised to find them loaded with those transporters," Rensing said. "What do they use them for? Nobody knows, but apparently the ability to regulate copper levels very precisely must be important to them."
"Metals and their availability to microbes have a huge impact on global cycles including the climate," Rensing said. "Exploring the roles that metals play in those processes and how bacteria transfer them into and out of their cells is a new frontier."
More information: Switch or Funnel: How RND-Type Transport Systems Control Periplasmic Metal Homeostasis, The Journal of Bacteriology, May 2011,p. 2381-2387, Vol. 193, No. 10 doi:10.1128/JB.01323-10
Abstract
Bacteria have evolved several transport mechanisms to maintain metal homeostasis and to detoxify the cell. One mechanism involves an RND (resistance-nodulation-cell division protein family)-driven tripartite protein complex to extrude a variety of toxic substrates to the extracellular milieu. These efflux systems are comprised of a central RND proton-substrate antiporter, a membrane fusion protein, and an outer membrane factor. The mechanism of substrate binding and subsequent efflux has yet to be elucidated. However, the resolution of recent protein crystal structures and genetic analyses of the components of the heavy-metal efflux family of RND proteins have allowed the developments of proposals for a substrate transport pathway. Here two models of substrate extrusion through RND protein complexes of the heavy-metal efflux protein family are described. The funnel model involves the shuttling of periplasmic substrate from the membrane fusion protein to the RND transporter and further on through the outer membrane factor to the extracellular space. Conversely, the switch model requires substrate binding to the membrane fusion protein, inducing a conformational change and creating an open-access state of the tripartite protein complex. The extrusion of periplasmic substrate bypasses the membrane fusion protein, enters the RND-transporter directly via its substrate-binding site, and is ultimately eliminated through the outer membrane channel. Evidence for and against the two models is described, and we propose that current data favor the switch model.
Provided by University of Arizona