Multi-omics strategy gives systems-level insight toward Salmonella pathogenesis
Researchers from Pacific Northwest National Laboratory and Oregon Health & Science University combined extensive ‘omics measurements with a novel computational network analysis approach to better understand the complex regulatory interactions in the pathogen Salmonella Typhimurium and identify virulence-related proteins that cause systemic infection in a host. Their results, which appeared in BMC Systems Biology, demonstrate the utility of this approach to infectious disease research and offer new biological insights that may lead to new prevention and treatment strategies.
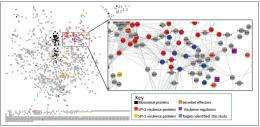
Parallel transcriptomics and proteomics measurements of Salmonella wild-type and 14 mutant strains, each lacking a specific regulatory protein required for systemic infection, provided data for a systems approach to network analysis. From the resulting network predictions, the research team discovered proteins not previously known to be required for virulence. Additional cellular analysis revealed a subset of these bacterial proteins that were secreted into the host cytoplasm independent of the known Salmonella mechanisms for transporting virulence proteins into host cells.
"Of particular interest, we identified two new virulence proteins that revealed a new class of secreted virulence proteins and that we are currently investigating," said PNNL scientist Dr. Charles Ansong, one of the investigators.
Salmonella is a leading cause of food-borne illness and is the culprit bacterium in high-profile food recalls, such as peanut butter in 2008/2009 and the current one of ground turkey that has led to 77 illnesses and one fatality reported in the U.S. to date. Systemic bacterial infections such as those caused by Salmonella are highly regulated and complex processes that include sophisticated offensive and defensive strategies by both pathogen and host that are orchestrated by virulence factors. Identifying the proteins required for Salmonella virulence is important for developing approaches to effectively combat persistent and adapting pathogens.
The researchers used capillary liquid chromatography-mass spectrometry to extensively characterize proteins present in the Salmonella wild-type and mutant strains. Parallel whole-genome microarray analysis was used to determine the transcriptome in matched samples.
More information: Yoon H, C Ansong, JE McDermott, MA Gritsenko, RD Smith, F Heffron, and JN Adkins. 2011. "Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella." BMC Systems Biology 5:100. DOI: 10.1186/1752-0509-5-100
Provided by Pacific Northwest National Laboratory