March 12, 2012 feature
Modeling the miniscule: High-resolution design of nanoscale biomolecules
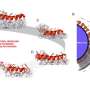
(PhysOrg.com) -- A key element of both biotechnology and nanotechnology is – perhaps unsurprisingly – computational modeling. Frequently, in silico nanostructure design and simulation precedes actual experimentation. Moreover, the ability to use modeling to predict biomolecular structure lays the foundation for the subsequent design of biomolecules. Historically, the problem has been that most modeling software presents a tradeoff between being general purpose (in being able to model systems at high/atomic resolution) but limited in scope (i.e., only explores a small fraction conformational space around the initial structure). Recently, however, Stanford University scientists have developed an algorithm – implemented in a modeling program known as MOSAICS (Methodologies for Optimization and SAmpling In Computational Studies) – that achieves nanoscale modeling at the resolution required without being limited by the scope/size dilemma. In addition, the researchers successfully modeled – and benchmarked the new computation modeling technique with – RNA-based nanostructures.
The research team – Adelene Y. L. Sim in the Department of Applied Physics, and Prof. Michael Levitt and Dr. Peter Minary in the Department of Structural Biology – faced a range of challenges in devising their unique algorithm. Speaking with PhysOrg, Minary and Sim describe those challenges. “Reducing dimensionality may eliminate physically relevant paths connecting conformational basins and therefore introducing artificial energy barriers that do not present obstacles in Cartesian space,” Minary tells PhysOrg. “In the present case, the major challenge was to develop an algorithm that supports degrees of freedom representing arbitrary collective rearrangements at all-atom resolution.”
Unfortunately, Minary notes, using these degrees of freedom, or DOFs, could break chain connectivity – and the corresponding conformational space is likely to be associated with extremely rough energy surface topology. “To overcome these limitations,” he adds, “less collective rearrangements need to be utilized only at a necessary extent so that rearrangements along the more collective DOFs are optimally facilitated without significantly increasing the volume of the sampled conformational space.” In short, their major challenge was implementing a universal algorithm capable of exploring conformational space while allowing numerous sets of arbitrary and/or user-defined so-called natural DOFs.
The team addressed these issues, Minary says, by building on the preexisting high-level computational environment of the MOSAICS software package that enabled the use of arbitrary even chain breaking DOFs. “To further improve on this concept,” he adds, “a very flexible new interface had to be invented that welcomes users to define their own system specific DOFs. In addition, the interface also had to support the weighted superposition of arbitrary DOFs. Finally a universal algorithm that realizes the interaction of various sets of DOFs needed to be implemented.” By so doing, conformational paths along the most collective molecular rearrangements are augmented by the incorporation of progressively more detailed molecular flexibility without significantly altering the dimensionality problem, which is better quantified by the conformational volume to be sampled rather than the actual number of DOFs.
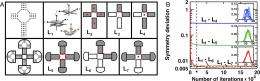
Other innovations are also in the works. “In the current paper we showed that our algorithm satisfies some necessary conditions of phase space, or detailed balance, preserving sampling not satisfied by any of the available algorithms used to model RNA systems,” Minary notes. “Further efforts are invested to fully satisfy microscopic reversibility.” Moreover, computational efficiency may be improved by using information on the collective nature of DOFs when updating atomic interactions, or by defining energy functional forms in terms of low dimensional analytical coordinates. Minary points out that sampling efficiency could also be improved if the current approach is combined with some advanced sampling algorithms based on multi-canonical sampling available in MOSAICS.
In addition, he continues, the movement of explicit water could be incorporated into the hierarchical moves so that effects of solvation can be more accurately evaluated – and testing the method with various implicit solvent representations may also be informative. “Finally,” he says, “we’re planning to introduce a more user friendly – possibly graphical – interface that would bridge the gap between algorithm developers and computational biologists, physicists and chemists who have great insight and intuitions about the natural DOFs of various molecular assemblies and complexes.” Altogether, all the above efforts, which would increase mathematical rigor, computational speed, solvent details and accessibility to users, could further extend the boundaries of applications beyond the current systems being considered.
In the meantime, while developing all the necessary algorithms discussed above, the team plans to continue extending the range of target applications. “Besides modeling the structure of chromatin,” Minary illustrates, “we’d like to revisit questions in DNA nanotechnology.” Furthermore, the use of a refinement method other than Cryo-EM (Cryo-Electron Microscopy, a form of transmission electron microscopy where samples are studied at cryogenic temperatures, and which the team is already pursuing) is also planned.
“We intend to extend our work to extensively explore RNA junction flexibility,” adds Sim, “and are also currently looking into using our technique in RNA structure prediction of large RNA systems.” In terms of applications, Sim continues, “in medicine it’s vital to understand the flexibility, stability, shape and possible distortions of nanostructures to better evaluate the nanostructure quality. These properties could play crucial roles in dictating cellular internalization and/or toxicity of nanostructures.”
Sim points out that with their efficient modeling tool, although still dependent on the quality of the force field used, the team is now more capable of studying these properties in silico. “Additionally,” Sim notes, “we’re looking into optimization in sequence- and structure-space simultaneously by having sequence as an additional degree of freedom.” A possible application is the sequence design of silencing RNA, or siRNA.
Looking further afield, Minary tells PhysOrg, there are other technologies and applications that might benefit from their findings. “Since proper sampling and exploration of the conformational space is a basic tool used in various technologies and applications, the method could be used in design, homology modeling and various new applications such as modeling collective rearrangements in trans-membrane proteins, designing new nucleic acid nanostructures, modeling large protein-nucleic acid assemblies, such a the ribosome, and the in silico study of chromatin remodeling. In addition,” he adds, “we’d like to aid the refinement and interpretation of experimental techniques.” Specifically, building on former efforts to refine Cryo-EM data, they’d like to develop tools to analyze NMR, FRET, SAXS, X-Ray, and footprinting experiments in order to generate conformational ensembles that satisfy experimental constraints.
Finally, Minary points out that the algorithm they developed is very general in nature and could be also utilized in other disciplines that involve state spaces with a large number of variables that are changing in a correlated manner. “In particular,” he concludes, “the basic idea could be used but not limited to sampling the space of possible networks, as in systems biology applications, or stock market variables.”
More information: Modeling and design by hierarchical natural moves, PNAS February 21, 2012 vol. 109 no. 8 2890-2895, doi: 10.1073/pnas.1119918109
Related:
The effects of polymeric nanostructure shape on drug delivery, Advanced Drug Delivery Reviews, Volume 63, Issues 14–15, November 2011, Pages 1228–1246, doi: 10.1016/j.addr.2011.06.016
Square-Shaped RNA Particles from Different RNA Folds, Nano Letters, February 24, 2009 (Web), 9 (3), 1270–1277, doi: 10.1021/nl900261h
Copyright 2012 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.