Shedding light on photosynthesis

(PhysOrg.com) -- Imagine being able to monitor protein expression levels in a cell as they change over time and in response to external stimuli. That is just what researchers did when they studied the photosynthetic blue-green algae, Cyanothece 51142, as part of the Membrane Biology Scientific Grand Challenge at EMSL. The Challenge's team members from Pacific Northwest National Laboratory and Washington University in St. Louis measured dynamic changes in protein expression within Cyanothece when it performed photosynthesis.
This technique of dynamic proteome analysis goes a step beyond static, traditional proteomics studies, provides greater insight into how proteins are expressed as a function of time, and will enable future similar studies of not only other cyanobacteria but other species of bacteria in general. Moreover, the team's results provide new knowledge about photosynthesis—nature's elegant way of manufacturing clean energy.
The scientists first cultured Cyanothece cells for several days under normal growth conditions, then introduced media containing carbon-13 and nitrogen-15-labeled leucine, an amino acid easily taken up by the cells for efficient synthesis of new proteins. They stimulated photosynthesis by subjecting the cultures to strong light. They then harvested cell samples over 48 hours, collecting both unlabeled and labeled proteins expressed at each time point.
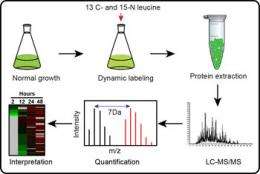
"This is an efficient technique for real-time monitoring of new protein synthesis because it allows us to collect protein samples at multiple time points and quantitatively measure newly synthesized proteins that result from changes in environmental or cultural growth conditions, such as high light intensity," said Dr. Uma Aryal, PNNL postdoctoral researcher and lead author of the paper.
The information gained from such studies using this technique would be highly useful to optimize Cyanothece and cyanobacteria for specialized environments or energy production, such as increasing CO2 fixation efficiency.
Each sample was analyzed with EMSL's liquid chromatography-tandem mass spectrometry and bioinformatics resources. By comparing the relative isotope abundances of each labeled and unlabeled peptide, the researchers identified 414 different proteins with significant changes in abundance over the 48-hour duration of the experiment.
Researchers will apply this metabolic labeling strategy to other cyanobacterial systems for quantitative measurement of their cellular proteomes. The eventual goal is to increase the rate at which cyanobacteria are capable of taking up carbon dioxide.
More information: Aryal UK, et al. 2012. "Dynamic Proteome Analysis of Cyanothece sp. ATCC 51142 Under Constant Light." Journal of Proteome Research 11(2):609-619.
Journal information: Journal of Proteome Research
Provided by Pacific Northwest National Laboratory