Evolution of an imprinted domain in mammals
The normal human genome contains 46 chromosomes: 23 from the mother and 23 from the father. Thus, you have two copies of every gene (excluding some irregularity in the pair of sex chromosomes). In general, which parent contributes a chromosome has no effect on the expression of the genes found on it. Exceptions to this rule are caused by “genomic imprinting”—modification of DNA, which means that gene expression is influenced by which parent the gene came from. A new paper published this week in the open-access journal PLoS Biology investigates the evolution of genomic imprinting in a specific region of the mammalian genome.
The work, by Anne Ferguson-Smith and colleagues in the UK and Australia, shows that different regions became imprinted at different times during mammalian evolution.
Genomic imprinting is hypothesized to have evolved because of the conflicting interests of mother and father relating to offspring development. In some conditions where two parents contribute to producing young together, each parent benefits evolutionarily by coercing the other into investing more in the baby—because the investment will benefit their own genes (in the form of their child) and cost an individual that is genetically unrelated—the mate.
Therefore, genes in the father may benefit from producing a placenta that demands a lot of maternal resources, and thus there would be a selection pressure to modify sperm—but not eggs—so that the genes they carry are expressed in a way that builds a demanding placenta. Indeed, in mammals, imprinting seems to have arisen in line with the evolution of the placenta and the new work by Ferguson-Smith et al. supports this.
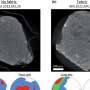
There are three existing lineages of mammals: placental mammals (including humans), marsupial mammals (which have a simpler form of placenta, and include kangaroos, koala bears, etc.), and monotremes (egg laying mammals, e.g. the duck-billed platypus). When comparing these three mammalian groups at a specific region of the genome (called Dlk1-Dio3), Ferguson-Smith et al. found that imprinting occurred only in placental mammals.
This finding contrasts previous work, which has found regions imprinted in both marsupials and placentals, but not in monotremes. Thus, together with previous work, Ferguson-Smith et al. have shown that imprinting of the mammalian genome occurred gradually; some genes becoming differentially expressed before the marsupial-placental common ancestor and others afterwards. That different regions changed at different times suggests that these changes were in response to selection pressures and therefore are adaptive—beneficial to survival/reproductive fitness rather than a by-product of another process.
Interestingly, the genetic comparisons Ferguson-Smith and colleagues have made show that imprinting correlates with highly repetitive regions of DNA. In marsupials, the Dlk1-Dio3 region is double the length found in placental mammals, due to random insertion of non-coding DNA, whereas in the different placental lineages, the region has very little non-coding sequence.
Citation: Edwards CA, Mungall AJ, Matthews L, Ryder E, Gray DJ, et al. (2008) The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol 6(6): e135. doi:10.1371/journal.pbio.0060135 (biology.plosjournals.org/perls … journal.pbio.0060135)
Source: Public Library of Science