E. coli discovery could lead to new antibacterial target
Northeastern University scientists have discovered a new and unique DNA binding property of a protein in E. coli. Penny J. Beuning, Assistant Professor in the Department of Chemistry and Chemical Biology, spent the last two years researching double and single-stranded DNA binding of E. coli DNA polymerase III alpha protein and notes that her findings have potential for developing a new antibacterial target.
Beuning's results have recently been published in ACS Chemical Biology in an article titled "Distinct Double- and Single-Stranded DNA Binding of E. coli Replicative DNA Polymerase III Alpha Subunit".
This work represents the collaborative effort of the Northeastern laboratories of Beuning and Mark C. Williams, Associate Professor of Physics, and involved researchers Micah J. McCauley, Leila Shokri, and Jana Sefcikova from both laboratories. Additionally, Česlovas Venclovas, of the Institute of Biotechnology in Lithuania, provided computational modeling expertise to the project.
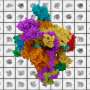
The project took advantage of the single-molecule expertise in the Williams laboratory and used a series of optical tweezers experiments to find that the DNA polymerase subunit of the 10-subunit bacterial replicative DNA enzyme has affinity for both double and single-stranded DNA in distinct subdomains of the protein.
DNA polymerase III is responsible for copying the entire genome of E. coli every time a cell divides. The alpha subunit is the enzyme that actually copies the DNA, and that activity is well-known. However, there are additional parts of the protein that were not characterized and that the researches suspected had DNA binding activity. The researchers first confirmed that the protein binds both double- and single-stranded DNA. Using protein engineering methods to isolate protein domains, they were able to localize the two different DNA binding activities to two different domains of the protein.
"The single-stranded DNA binding component appears to be passive, because the protein does not assist in melting but instead binds to single-stranded regions which are already separated by force," said Beuning. "Detecting this kind of binding would be difficult or impossible using traditional methods of assaying DNA binding activity."
The researchers' results demonstrated that single-stranded DNA binding is localized to an OB-fold domain while a tandem, helix-hairpin-helix motif contributes significantly to double-stranded DNA binding. Single-stranded DNA binding by the subunit occurs only after single-stranded DNA has been fully melted by force. This unusual behavior, noted Beuning, may be functionally important as single-stranded DNA binding will likely occur only after other replication processes create single-stranded DNA.
“It is crucial to understand how these kinds of massive biological machines function in the cellular environment in order to fully exploit their potential as drug targets,” added Beuning.
Provided by Northeastern University