April 15, 2009 feature
Researchers Build World's Largest Disease Association Network
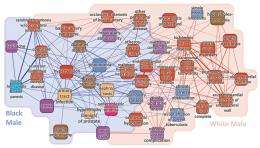
(PhysOrg.com) -- If you suffer from hypertension, how much does your risk for developing diabetes or other illnesses increase? Medical experts have long known that many diseases are related to one another, even to the point that there are often no clear boundaries between different diseases. Such correlations occur because diseases can have multiple causes, and also because multiple diseases can have the same cause (for example, the same gene or protein pathway).
Understanding the connections among diseases is extremely challenging, yet it could lead to better healthcare in the form of earlier diagnoses and treatments. In a recent study, researchers from Northeastern University in Boston, Massachusetts, and Harvard University in Cambridge, Massachusetts, have analyzed data from 32 million individuals’ insurance claims to build the largest ever disease association network. Their Phenotypic Disease Network (PDN) identifies correlations among more than 10,000 diseases. The researchers have made the data freely available to other researchers, doctors, and patients at hudine.neu.edu , where it can be downloaded in bulk or viewed in the form of graphs and tables that summarize the associations between a particular disease and all other diseases. The study is published in a recent issue of PLoS Computational Biology.
“In the great scheme of things, we can think that there are two ways to understand the things that surround us,” lead author César Hidalgo of Harvard University told PhysOrg.com. “One is to try to understand what things are made of, i.e. take the objects that make up the world and break them up in order to study their components. The second way is to take the objects that make up the world and see how they relate to other objects, in what context they occur and how they are connected. Until now, medicine has concentrated most of its efforts in generating understanding by disaggregating their objects of study (diseases) into essential components (genes, proteins, pathways, organs). Here we show that it is in principle possible to characterize and understand diseases also by looking at their context, rather than their components.”
As the researchers explain, their data comes from 32 million Americans aged 65 or older enrolled in Medicare. The data involves 32 million claims from 13 million individuals from 1990 to 1993 (the remaining individuals were not hospitalized during this period). The researchers compiled the data in the form of ICD-9 classifications, which consisted of 17,000 different disease categories. While most of these diseases were rarely diagnosed, a few were diagnosed in a large fraction of the population.
The Phenotypic Disease Network consists of nodes (diseases) and links between diseases. As such, the PDN can be viewed as a “map” of illness progression in space, in which patients “jump” from one disease to another along the network’s links. When an individual is affected by two diseases, the diseases share a “comorbidity relationship.” To quantify this relationship, the researchers used two different metrics: relative risk and Φ (phi) correlation. Due to intrinsic bias, the relative risk metric is more sensitive to rare diseases, while the phi correlation network is more sensitive to common diseases, and so each metric excels at a different scale.
The researchers found that the structure of the disease networks could increase their understanding of illness progression in several ways. For instance, they found that patients are more likely to develop diseases close in the network to diseases they already have. In a sense, the development of a patient’s illness can be thought of as a spreading process over the network.
In addition, diseases that are highly connected tend to be preceded by less connected diseases, and highly connected diseases are associated with higher degrees of mortality. In this way, a patient’s location in the network can serve as a predictor of the number of years he or she is expected to live. The researchers also found that an individual’s gender and ethnicity altered their personal disease network.
Overall, the study shows that a network can serve as a valuable representation for disease progression, and offers the potential to help researchers better understand the origin and evolution of human diseases. By combining this kind of phenotypic information with genetic and proteomic information, researchers could have a broader perspective of illness progression and comorbidity relationships. Also, as healthcare becomes more delocalized, a map like the PDN could also be an ideal way to represent medical records for healthcare workers.
“One possible application of medical health records is to compare the overall health status of different populations (e.g. cultural, geographical, etc.),” Hidalgo said. “Disease networks can inform us not only about the difference in prevalence between diseases, but also about differences in the strength of disease associations observed in different populations. In general, diseases have multiple causes and therefore differences in the associations between diseases are expected to be informative about the causes of diseases in different populations.”
He added that, in the future, doctors could use digital flip-charts to access the medical records of a patient, in which a patient's disease history would be represented by highlighted nodes in a disease network. Not only could the doctor see a list of the diseases that have previously affected that patient, but also other diseases that tend to co-occur with the patient’s diseases. Furthermore, the genes associated with those diseases could be only a click away.
“Disease networks could also be informative for drug companies that could be interested in new potential indications of their drugs,” Hidalgo said. “They could also be helpful for hospital design, as given limited budget constraints, hospitals may want to be designed to not leave out diseases that are strongly connected to other diseases they are already treating.”
More information: Hidalgo, César A.; Nicholas Blumm; Albert-László Barabási; Nicholas A. Christakis. “A Dynamic Network Approach for the Study of Human Phenotypes.” PLoS Computational Biology, April 2009, Volume 5, Issue 4, e1000353. www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1000353
Copyright 2009 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.















