Engineering bacterial cells
(PhysOrg.com) -- Two teams of Oxford University researchers led by Professors Judith Armitage and David Stuart have made the first steps towards being able to engineer a bacterial cell that can sense and respond to novel environmental cues. The groups demonstrated that it should be possible to design synthetic signalling circuits inside a cell, ultimately enabling the development of new biosensors.
The ability of bacteria to respond to environmental cues is a universal feature of living cells. Evolution has created a vast array of signalling mechanisms that enable cells to react in many ways to changes in their surroundings. One of the most important of these is a two-component signalling circuit which is widely used by bacteria. It comprises a protein kinase, which acts as the sensor, and its partner protein known as a response regulator.
Some species of bacteria have over 150 different sensor-response regulator pairs in a single cell, so the specificity between these pairs has to be tightly controlled to prevent ‘crossed wires’ between signalling pathways.
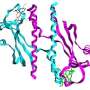
The two Oxford groups combined their expertise in structural biology and biochemistry to address where this specificity comes from. They used extremely bright pinpoints of light produced by the UK’s national synchrotron facility, Diamond Light Source in south Oxfordshire, to carry out X-ray crystallography, a method that allows visualization of proteins at an atomic level. Using this approach, the researchers solved the three-dimensional structure of one of these two-component complexes found in the bacterium Rhodobacter sphaeroides.
This complex is crucial for a process known as chemotaxis which controls the movement of a bacterium when it senses a chemical or nutrient gradient in its environment. Based on the crystallography results, the researchers were able to pinpoint the specific amino acids that are required for this molecular recognition. They found that one amino acid on the response regulator pointed out like a finger towards a pocket on the sensor, enabling the two proteins to fit snugly together.
By introducing this finger into other response regulator proteins, which do not normally partner this specific sensor, they were able to change their specificity and re-engineer the chemotaxis pathway.
This is the first time that researchers have re-designed the intracellular part of the chemotaxis circuitry. This re-engineering paves the way for producing custom-designed circuits for applications in systems biology.
“This is a significant step on the road to identifying the critical amino acid interface that allows discrimination between apparently related proteins and their partners, and a step along the road to rational design of protein signalling networks,” Professor Judith Armitage said. “The aim is to understand the system so well that you're able to change it in any way you like”, says Christian Bell, the postgraduate student who worked on the project in both labs. “The dream will be a synthetic cell that does exactly what you want.”
More information: Bell CH et al. (2010). Using Structural Information to Change the Phosphotransfer Specificity of a Two-component Chemotaxis Signalling Complex. PloS Biology 8(2): e1000306. doi:10.1371/journal.pbio.1000306
Provided by Diamond Light Source