Rapid, Inexpensive DNA Sequencing Moves Closer to Reality
As efforts such as The Cancer Genome Atlas and others generate vast quantities of information about the genetic makeup of different types of cancer, it is becoming increasingly clear that such information has great potential for determining which anticancer drugs should be used to treat a specific patient. However, realizing that potential will require not only that cancer researchers uncover the links between specific gene changes in a given tumor and that tumor's response to a specific drug therapy, but that technologists develop faster methods of detecting specific mutations that would be economical to use on individual patients.
A technological breakthrough to address this latter issue may be at hand thanks to recent work conducted by Amit Meller and his colleagues at Boston University. Reporting their work in the journal Nano Letters, these investigators described the use of electrically charged nanopores to detect specific genetic sequences as single DNA molecules pass through the pore. If further development proves successful, this method could yield a new approach to mutation detection that does not involve time-consuming and expensive amplification processes.
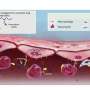
The investigators built their sequencing device by using a focused electron beam to drill a 4-5 nanometer diameter hole in a silicon nitride membrane. The membrane is then placed between two small fluid chambers and an electric field is applied across the membrane using a pair of silver/silver chloride electrodes. This applied current causes individual DNA molecules to move through the pore, untwisting and unraveling as they enter the pore.
In order to identify a known genetic sequence, the investigators first treat a DNA sample with specific sequences of a DNA analog known as a peptide nucleic acid, or PNA, that will bind to the proper complementary DNA sequence of interest. When the matched DNA-PNA sequence passes through the pore, it produces a marked change in the electrical current passing between the two electrodes, a change that the investigators demonstrated is easily distinguished from unaltered double-stranded DNA, that is, DNA not duplexed with the PNA probe. The device is capable of analyzing one DNA molecule per second.
This work is detailed in a paper titled "Nanopore Based Sequence Specific Detection of Duplex DNA for Genomic Profiling." An abstract of this paper is available at the journal's Web site.
Provided by National Cancer Institute