Refined tools help pinpoint disease-causing genes
In findings that may speed the search for disease-causing genes, a new study challenges the prevailing view that common diseases are usually caused by common gene variants (mutations). Instead, say genetics researchers, the culprits may be numerous rare variants, located in DNA sequences farther away from the original "hot spots" than scientists have been accustomed to look.
Using an approach that detects rare but powerful causal gene variants, the researchers say they have accounted for a significant proportion of the "missing heritability" problem - the disappointing fact that, to date, conventional gene-hunting studies have often failed to identify, when searching for gene variants, variants that cause a large proportion of common diseases, such as heart disease, cancers and diabetes.
The new approach draws on existing data from genome-wise association studies (GWAS) that have already been performed, reanalyzing the data to pinpoint causal variants that have not been identified previously.
Furthermore, the technique may allow researchers to identify individuals whose DNA is more likely to carry specific mutations in the causal genes. "Our approach draws us closer to the goal of personalized medicine, in which treatment will be tailored to an individual's genetic profile," said study leader Hakon Hakonarson, M.D., Ph.D., director of the Center for Applied Genomics at The Children's Hospital of Philadelphia. "When we can say that a specific gene mutation causes a patient's disease, we have more meaningful diagnostic results. Identifying causal variants in disease genes provides an opportunity to develop drugs to rectify the biological consequences of these mutated genes."
Hakonarson and colleagues, including first author Kai Wang, Ph.D., of Children's Hospital, collaborated with David B. Goldstein, Ph.D., director of the Center for Human Genome Variation at Duke University. The study appears online today in The American Journal of Human Genetics.
Earlier this year, Goldstein led a study, in collaboration with Hakonarson and Wang, that presented a model of this approach, performing computer simulations based on GWAS data. The current study strengthens the model by analyzing real DNA sequencing data from two well-known diseases—Crohn disease and genetic hearing loss.
A GWAS uses gene chips in automated systems that analyze about 500,000 to one million sites where single-letter differences in DNA tend to occur; these differences are called single-nucleotide polymorphisms, or SNPs. In using these SNP chips over the past decade in comparing DNA samples between healthy subjects and patients, scientists have identified thousands of SNPs that associate with common complex diseases. However, geneticists believe that the SNPs investigated by the gene chips do not themselves cause a disease, but instead serve as a "tag," a marker linked to the actual causal mutations that may reside in a nearby region.
After a GWAS finds SNPs linked to a disease, researchers then perform a "fine-mapping" study by additional genotyping--sequencing of the gene regions near the SNP signal, to uncover an altered gene that harbors a mutation responsible for the disease. Most of the results, said Hakonarson, have been unimpressive, yielding causal variants with very small effects. "These efforts have not identified 'smoking gun' mutations that cause a disease," he added.
The current study uses different assumptions. Instead of inferring the presence of a nearby common disease-causing gene linked to a given SNP, the researchers propose that numerous rare causal variants may separately "hitchhike" on the same tag SNP, often from locations further away than those scrutinized in conventional fine-mapping approaches. Those more distant genetic factors are typically overlooked in the conventional GWAS approach. By missing these actual causative gene variants, the conventional technique underestimates the strength of their effect on the disease.
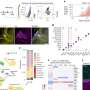
By applying their methods to real DNA samples from patients with genetic hearing loss, the researchers' approach helped them to select from GWAS data a subset of cases for sequencing analysis that were most likely to carry causative mutations. Sequencing the DNA in this subset, the study team found that the majority of those patients carried an actual mutation known to cause hearing loss. "Our technique suggests that when we do our resequencing follow-up studies, we can identify people who are much more likely to carry a causative gene," said Kai Wang, who analyzed the dataset. Hakonarson added, "We present a more efficient approach for mining GWAS data to find the actual causative gene variants that will have future utility in designing therapies."
More information: "Interpretation of Association Signals and Identification of Causal Variants from Genome-Wide Association Studies," The American Journal of Human Genetics, published online April 29, 2010. dx.doi.org/10.1016/j.ajhg.2010.04.003