Shining Light on Brain Tumours
(PhysOrg.com) -- The Canadian Cancer Society estimates that 2600 new cases of brain cancer will be diagnosed in 2010. Glioblastoma multiforme (GBM) is the most aggressive and malignant form of brain cancer, and attempts to successfully treat GBM tumours depends on identifying tumour cells - both when detecting cancer in biopsies and when ensuring that all of the cancer has been removed after surgery. With the help of the Canadian Light Source, a team led by Dr. Kaiser Ali, a pediatric oncologist in the Saskatchewan Cancer Agency's Cancer Research Unit and collaborators from the University of Saskatchewan, Saskatoon Health Region, CLS and the National Research Council have been able to identify a chemical signature unique to GBM tumour cells. The pilot study made the front cover of the July 2010 issue of the International Journal of Molecular Medicine.
“For me, the synchrotron is like the fingerprinting powder used by investigators,” says Dr. Ali. “Using the synchrotron’s infrared beamline is like dusting for fingerprints - we “dust” the tumour to detect its molecular fingerprint pattern. We can then ID the ‘perpetrator’ from its own unique fingerprint.”
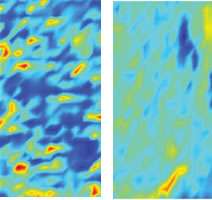
Dr. Ali and his team compared paired slices of cancerous and healthy brain tissue that had been removed from GBM patients. One of the specimens from each pair was stained and examined by a pathologist using a regular light microscope. The second specimen was then examined under the synchrotron’s infrared spectromicroscope, which is capable of detecting the tell-tale signatures of biomolecules such as proteins, carbohydrate and fat components inside individual cells and build a chemical map.
By overlaying a new kind of digital grid developed by the research team over the stained specimens and the synchrotron images, the position of cancer cells in the conventional slides were very closely correlated with their position on the synchrotron maps. The results were striking - the cancer cells contained far lower concentrations of lipids (fats), as well as different levels and ratios of proteins and carbohydrates compared with healthy brain tissue. The contrast is obvious in the computer-generated colour images featured on the journal’s cover, with the dark blue assigned to lipids that mottles the healthy tissue in almost regular globs, all but absent from the image taken from a tumour.
A larger study looking at GBM has just been completed, and analysis of the results are under way. Dr. Ali is optimistic that this synchrotron technique can be a powerful new addition to the arsenal in diagnosing cancer more accurately. He also sees a wide range of practical applications of the synchrotron’s capabilities, from detection and identification of tumours, to improved understanding of how cancers grow and spread, to treatment using synchrotron generated X-rays.
“I am a clinician-scientist who entered the door of synchrotron research with a sense of curiosity, worked at synchrotron experiments for over seven years, and now see myself as a kindergarten student - just starting to understand what it’s all about”.
More information: K. Ali, et al. 2010. Biomolecular diagnosis of glioblastoma multiforme using Synchrotron mid-infrared spectomicroscopy. International Journal of Molecular Medicine 26 pp. 11-16. DOI: 10.3892/ijmm_00000428