From predictions to reality: Genomics reveals microbe's metabolic potential
Knowing an organism's metabolism can give scientists essential insights into how the organism uses its resources. These insights can then enable them to tweak the metabolism to enhance the microbe's use of these resources in beneficial ways, such as to reduce contamination in soil or to produce biofuels or other desirable chemicals efficiently.
With the latter goal in mind, collaborators from Pacific Northwest National Laboratory (PNNL), Burnham Institute for Medical Research, University of Wisconsin-Madison, and Dartmouth College used computational and high-throughput experimental approaches to reconstruct the metabolic network of Shewanella oneidensis. This has enabled the team to develop a predictive understanding of the microbe's metabolism.
For years, Shewanella has been of great interest to the Department of Energy because of its ability to reduce metals and radionuclides by extracellular electron transfer. This also makes the metabolically versatile organism an ideal candidate for use in microbial fuel cells. In 2002, the strain S. oneidensis MR-1 was among the first bacteria to have its genome completely sequenced, providing an initial glimpse into the untapped metabolic potential of the organism and laying a foundation for the studies to follow.
"To optimize the electron transfer process and carbon metabolism so that we can direct Shewanella metabolism toward biofuel production, we need to be able to understand and predict the cell's metabolism at a system level. Modeling the cell metabolism in silico—by computer simulation—is the key to achieving this," said Dr. Grigoriy Pinchuk, lead author of a paper that appeared in PLoS Computational Biology.
"Shewanella cells obtain energy by oxidizing organic compounds and transferring electrons through a chain," Pinchuk added. "The cells use this energy for all kinds of functions including biosynthesis of new cell components. Optimal electron transport and carbon metabolism are important for the cell's well being. That's why genomic-scale modeling combined with experimental approaches is critical to obtain a deeper understanding of metabolism and refining the model so the final results better reflect the real world. Doing so will help us design processes by computer simulation and then implement them."
Shewanella is also an interesting candidate for biofuel production because of its ability to synthesize appreciable amounts of biofuel precursors such as lipids and fatty acids. Pinchuk said, "It seemed that if we could deeply understand the energetics and metabolism of Shewanella, we could harness this capability."
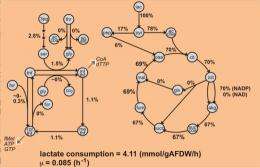
The research team built a genome-scale metabolic model and used it to analyze experimental data and predict cellular phenotypes. They compared predicted cellular biomass yields on different carbon sources in the presence of oxygen to experimental measurements. The results were used to refine metabolic fluxes for growth on lactate—a food and energy source for the bacterium. Analyses of S. oneidensis MR-1 metabolism revealed some of the reasons for a significant difference between experimental and predicted growth efficiency on lactate. The developed model provides a platform for a systematic assessment of Shewanella metabolism, which may be used for redesigning metabolic networks for chemical production.
The scientists are continuing to further the understanding of metabolic pathways and their performance at the whole cell level, and even in mixed cultures, to lead to fast and efficient biofuels processing. In addition, Shewanella was recently shown by PNNL colleagues at the University of Minnesota to produce a polyolefinic hydrocarbon that would provide a more direct route to biodiesel (see references).
More information: References:
Pinchuk GE, EA Hill, OV Geydebrekht, J De Ingeniis, X Zhang, A Osterman, JH Scott, SB Reed, MF Romine, A Konopka, AS Beliaev, JK Fredrickson, and JL Reed. 2010. "Constraint-Based Model of Shewanella oneidensis MR-1 Metabolism: a Tool for Data Analysis and Hypothesis Generation." PLoS Computational Biology 6(6):e100082. doi:10.1371/journal.pcbi.1000822
Sukovich DJ, JL Seffernick, JE Richman, KA Hunt, JA Gralnick, and LP Wackett. 2010. "Structure, Function, and Insights into the Biosynthesis of a Head-to-Head Hydrocarbon in Shewanella oneidensis Strain MR-1." Applied Environmental Microbiology 76(12):3842-3849. doi:10.1128/AEM.00433-10
Provided by Pacific Northwest National Laboratory