Studying cell signaling using single-molecule imaging
The ability to observe individual proteins as they react and combine provides remarkable insights into the complex world of cell signaling
Single-molecule imaging (SMI) is a powerful scientific tool for visualizing individual molecules. Among its wide range of potential uses, SMI is particularly promising for its ability to provide new insights into the molecular processes in living cells. With this technique, it is possible to count the number of proteins in a cell and observe their distribution, movement, reaction rates and even changes in conformation. Yasushi Sako, chief scientist in the Cellular Informatics Laboratory of the RIKEN Advanced Science Institute (ASI), is investigating the complex signaling mechanism in cells by studying the proteins involved, one by one, using SMI. His research has revealed that the reactions of individual proteins are more complex than previously thought, and that signaling is controlled by dynamic conformational changes and reaction memory.
First encounters with single-molecule imaging
Sako remembers vividly the excitement he felt when he succeeded in using SMI to observe the molecules in a cell for the first time. “I saw many small twinkling dots that moved randomly back and forth. Their movements were so mesmerizing and fascinating that I couldn’t stop looking at them.” Single-molecule imaging is performed using a fluorescence microscope to observe the trace fluorescence produced by individual proteins that have been labeled with green fluorescent protein (GFP) by gene transfer or chemical reaction. The method is so effective that it allows the number, distribution and movement of proteins to be observed—it can even be used to trace which protein binds to which, and the time required for a certain reaction.
Single-molecule imaging was developed in 1995 by a group of researchers from Osaka University and other institutions. They succeeded in viewing the movement and chemical reaction of a molecule in aqueous solution in a test tube, for example, using myosin isolated from a cell. Myosin is a motor protein responsible for muscle movement. What happens in a test tube, however, may not necessarily happen in a living cell. Researchers around the world therefore started to compete to develop an SMI technique that could be used to observe the behavior of proteins within a living cell. Sako was not one of those researchers. “My research interest back then was cell membrane proteins. The labels I used were several tens to several hundreds of nanometers in size, much larger than the fluorescent proteins used for SMI. As we already had a technique to view the movement of a labeled molecule in a cell membrane protein in living cells, we did not need SMI.”
In 1997, Sako moved to Osaka University. “In order to continue the research I’d been doing at my previous laboratory, where I had used a two-photon excitation fluorescence microscope, I assembled a new device. But I could not see what I wanted, no matter what adjustments I made. It was then that I wondered if SMI might work—the idea just sprang to mind.”
At the time, Sako was working in the laboratory of Toshio Yanagida, one of the leading researchers involved in the original development of SMI. “There were researchers at the laboratory who were developing a new device for SMI, but none were successful. I attempted to make a device by copying a conventional instrument, and using that microscope I was lucky enough to be able to see molecules in a living cell.” Looking back at his unexpected success in 1998, he says, “I used what I had at that time. I had no preconceived ideas.” In 2000, he published a paper on the technique, and that is when his relationship with SMI began.
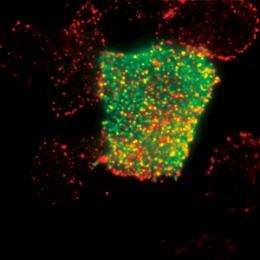
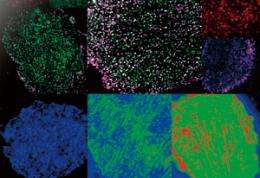
Understanding the response network of signaling proteins
Sako set up his Cellular Informatics Laboratory at the RIKEN ASI in 2006. “I wanted to know how the fate of cells is determined.” Cells may have a variety of fates: they may divide and increase in number, differentiate into different kinds of cells, or die (apoptosis). “Determination of the fate of a cell starts when a protein called a signaling molecule binds to a receptor embedded in the cell membrane,” says Sako. When bound by a signaling molecule, the receptor is activated and information is transmitted into the cell. The information is then conveyed from one protein to another within the cell through repeated binding, dissociation and migration until it eventually reaches the cell nucleus, where it induces the expression of a specific gene. This gene triggers various cellular responses, including proliferation, growth inhibition, differentiation, apoptosis, and oncogenic transformation.
There are many kinds of signaling molecules that can determine the fate of a cell. Among them, Sako is particularly interested in epidermal growth factor (EGF), which stimulates cell proliferation and movement. The response network of signaling proteins induced by EGF has been studied in detail and more than 100 proteins associated with the network have been isolated. “Based on the results of previous studies, we can draw a rough schematic of the network to show what elements are involved and how they are interconnected. But merely drawing a schematic does not mean we understand the response network. We need to observe the individual proteins that make up the response network in living cells directly to obtain quantitative information such as the number of proteins that elicit a response, their concentration and migration speed. Single-molecule imaging allows us to do this.”
Epidermal growth factor receptor undergoes dynamic conformational change
A receptor is thought to be activated when bound by a signaling molecule, but the epidermal growth factor receptor (EGFR) is not activated merely by the binding of EGF. The receptors are activated only when two EGFRs join together to form a dimer, and then only after EGF binds to both sides of the dimer. The mechanism of activation had not been fully elucidated until Sako decided to use SMI to study it in detail. There are about 50,000 EGFRs on the surface of a cell. The first thing he studied was the number of EGFs that need to be bound to EGFRs to induce a response. “I found that a response occurred when about 300 EGFs had bound to about 50,000 EGFRs. I counted the fluorescent dots one by one. Single-molecule imaging requires considerable patience!”
Those 300 EGFs on 50,000 EGFRs represent less than one percent. It is known that one to two percent of EGFRs form dimers, so how do so few EGFs find the dimers to bind to? “I found that EGFR uses a very sophisticated mechanism. A conformational change occurs in EGFR after dimerization, which makes it 100 times easier for EGF to bind to the dimerized EGFR than to the monomer EGFR. When EGF binds to one side of the dimer, the conformational change occurs again, which makes it another 10 times easier for EGF to bind to the other side of the dimer (Fig. 3). EGFR undergoes conformational change upon dimerization, which enables it to bind to EGF effectively and start the signaling process.”
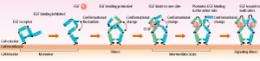
However, Sako stresses that the conformational change in EGFR is still speculative. “We need to investigate if such conformational change actually occurs. As it is possible to study conformational change through detailed observation of the fluorescence emitted from fluorescent proteins, we are now working on this study.”
Reaction memory in proteins
When EGFRs in a cell membrane are activated, phosphate binds to the intracellular part of the receptors. A protein that recognizes a phosphorylation site is then attracted to the site. There are more than ten different kinds of such proteins. Sako’s Cellular Informatics Laboratory is particularly interested in a protein called Grb2.
Upon recognition of the phosphorylation site of EGFR, Grb2 is attracted to the cell membrane. After a certain time, the Grb2 bound to the site will dissociate. The process up to this point is already well understood. In a study of the reaction rates of EGFR and Grb2 using SMI, however, researchers in Sako’s laboratory made a surprising discovery. “As the concentration of the binding protein increases, the reaction rate generally increases proportionally. However, even after increasing the concentration of Grb2 by ten times, we found that the reaction rate only increased three times. We consider that reaction memory may be involved in the reaction between EGFR and Grb2,” says Sako.
What is reaction memory? Upon binding to Grb2, EGFR undergoes conformational change. However, even after Grb2 has dissociated from EGFR, the receptor retains its bound conformation, returning to its original state only after a certain period of time. “EGFR, after completing the reaction, keeps a memory of the Grb2 bound to it. This is called reaction memory. Until the EGFR returns to its original unbound conformation, it is difficult for Grb2 to bind to it.” This explains why the reaction rate only increased three times despite the concentration of Grb2 being increased ten times. “It is possible in theory to use reaction memory to control the reaction rate in such a way that it remains unchanged even when the intracellular concentration of Grb2 changes. We are not sure yet if reaction memory is used for this purpose, and we will look at this issue in future research.”
Elucidating the complexity of protein reactions
The cellular responses induced as a result of signaling vary widely. For example, the binding of EGF to the receptor does not necessarily lead to cell proliferation; it may sometimes result in differentiation and, at other times, cell death. It has generally been understood that the diversity in cellular responses stems from the complexity of the response network of signaling proteins, whereas the reactions of individual proteins have been assumed to be very simple, involving just binding and unbinding. However, researchers are having to correct this long-held belief because of the discoveries Sako and his laboratory researchers have made using SMI. “It is true that the network is complex, but the reactions of individual proteins are also very complex. We have revealed that proteins control signaling by dynamic conformational change and reaction memory.”
The reactions of EGF, EGFR and Grb2 are just a small part of the beginning of the response network of signaling proteins. Is it possible to understand the entire network, which involves as many as 100 different kinds of proteins? “It may be difficult to observe all of the proteins that are associated with signaling using SMI. But I think, with the help of mathematical science, we can understand much of the network,” says Sako. The key reactions will be observed in detail and, based on the results, a model will be developed through computer simulations to better understand the entire network. “In our research, we collaborate with groups of researchers in mathematical biology and computational biology both within RIKEN and with other institutions.”
What does it mean to be alive?
Japanese researchers are pioneers of SMI, and yet Japan lags far behind the US in terms of the number of researchers and budget allocated to research in this field. “But we’re still far ahead in producing creative ideas and performing quality research.” For example, Sako is particularly interested in fluctuation. “This is a subject that Japanese researchers are good at. Researchers in Europe and the US prefer subjects that can be explained in a more clear-cut manner, so they pay little attention to fluctuation.” Cells are always exposed to fluctuation. The conformation of a protein, for example, is variously affected by thermal fluctuations, and there are also number fluctuations. “While cells have a mechanism to correct fluctuations, they also make good use of them. I want to demonstrate specific examples of how they use fluctuation in signaling and in determining their fate.”
Sako is philosophical about his research. “A protein is not defined as being alive, but a cell is alive. Something that is alive is generated from something that is not alive. Where is the boundary between them?” We still have no answer to this question. “When the border between the protein layer and the cell layer is removed, we may find the answer to what it means to ‘be alive’.”
Provided by RIKEN