Identifying enzymes to explode superbugs
With the worrying rise of antibiotic-resistant superbugs like MRSA, scientists from a wide range of disciplines are teaming up to identify alternative therapies to keep them at bay.
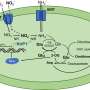
One long-considered solution is the use of lytic enzymes which attack bacteria by piercing their cell walls. Lytic enzymes are proteins that are naturally present in viruses, bacteria and in body fluids such as tears, saliva and mucus. However, until now, largely ad-hoc methods have been used to calculate the enzymes' killing abilities.
New research published tomorrow, Monday 4 October, in IOP Publishing's Physical Biology, shows how a group of US researchers have developed a pioneering method that can identify lytic enzymes for optimum bacteria killing characteristics.
In 1923, five years before discovering penicillin and laying the path for the development of antibiotics, Alexander Fleming had already noticed that a substance in mucus samples, lytic enzymes, could kill bacteria.
However, the success of antibiotics left the development of this finding in the shadows.
With the rise of antibiotic resistant superbugs, partially a result of antibiotics being a 'one-size-fits-all' therapy, Fleming's early discovery has been reinvigorated and lytic enzymes are back in the spotlight. Encouragingly, most lytic enzymes kill only a limited range of bacteria, unlike antibiotics, which allows researchers to target superbugs while potentially leaving beneficial bacteria intact.
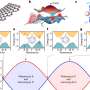
To identify the bacteria-killing characteristics of lytic enzymes Joshua Weitz and Gabriel Mitchell, quantitative biologists at the Georgia Institute of Technology, teamed up with Daniel Nelson, a biochemist from the University of Maryland, to identify, on a microscopic scale, the rate at which these enzymes pierce cell walls leading to bacterial death.
The piercing of cell walls can be fatal to bacteria because of a bacterium's internal pressure; the piercing is analogous to removing the wire on a shaken-up bottle of champagne.
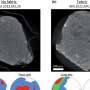
The researchers write, "While lytic enzymes and their associated antimicrobial activity have been studied for decades, their use as therapeutics has only recently been investigated in detail. We measured the amount of light passing through a bacterial solution, in much the same way as astrophysicists use light measurements for far-away galaxies: to infer processes at a far different scale based on interpreting the information contained in the light coming from them."
By measuring how lytic enzymes chemically clear a cloudy solution of living bacteria, the team was able to predict the cell level processes underlying bacterial death. In doing so, the researchers used the mathematical theory of inverse problems to overcome technical challenges in quantifying, for the first time, the microscopic killing properties of lytic enzymes.
The team was also able to estimate the extent to which genetically identical bacteria may be differentially susceptible to death via lytic enzymes.
"We believe we have taken the first step down a road which will allow us to identify more enzymes, choose those with the best activity, and engineer even higher activity, to develop an effective therapy against a wide range of dangerous superbugs."
The researchers hope their quest will result in "push button technology" to hasten the development of engineered enzymes for clinical use.
More information: iopscience.iop.org/1478-3975/7/4/046002
Provided by Institute of Physics