March 16, 2012 feature
New insights into ancient life: Chromosome segregation in Archaea
(PhysOrg.com) -- The effort to classify life into various groups has been a bumpy ride. Prior to the 1900s, living things were usually pegged as either plants or animals – period. By the middle of the 20th century, however, it was asserted that this scheme did not adequately represent fungi, bacteria and protists, leading to a five-group classification – Monera (bacteria), Protista, Fungi, Plantae, and Animalia. At roughly the same time, however, a fundamental distinction between prokaryotic bacteria and the four eukaryotic kingdoms (plants, animals, fungi, and protists) based on nuclei, cytoskeleton, internal membranes, and other shared eukaryote characteristics – for example, unlike eukaryotes, their genetic material is not wrapped by a membrane into a separate compartment – was acknowledged, resulting in a different system – and considerable confusion. Then, things changed anew when an entirely new prokaryotic group – the so-called third domain of life, living in high temperatures and producing methane – was discovered in the late 1970s.
Initially termed Archaebacteria (due to their typical but not universal morphological similarity to bacteria), the Archaea – found in a wider range of extreme as well as surprisingly mundane environments (such as bovine intestines) than first thought – are biochemically and genetically distinct from bacteria and eukaryotes, resulting in the current three-kingdom system of Archaea, Bacteria and Eukaryota. (Note that viruses are not organisms as defined by these three groups, and so constitute a fourth biological group.) Being hard to culture, little has been known about the genetic process by which they undergo chromosome segregation – a crucial step in species survival in which chromosomes pair off with their similar chromosomes, thereby ensuring that genetic material is accurately distributed to the next generation. Recently, however, scientists at the University of York and the Max Planck Institute in Marburg found that the archaeon Sulfolobus solfataricus uses a hybrid segrosome consisting of two proteins known as SegA and SegB.
The research team, led by Daniela Barillà in the Department of Biology at the University of York, and including lead author Anne K. Kalliomaa-Sanford and other researchers, faced several challenges in studying Archaea. “When we started this project,” Barillà tells PhysOrg, “all we knew was that the chromosome of Sulfolobus solfataricus harbored a gene, sso0034, related to bacterial genes involved in genome segregation. By inspecting the nearby DNA region, we noticed that the gene was followed by a short gene of unknown function, named sso0035. The intriguing thing was that the two genes were partially overlapping and, specifically, the end of sso0034 overlapped with the start of sso0035.” To the scientists, this architecture suggested that the two genes encoded proteins involved in the same biological process – meaning that one of their major challenges was to discover the function of the two proteins starting from scratch. “It’s like being at a crime scene,” says Barillà. “You have a few clues and you build on them to construct a jigsaw that reveals the final picture.”
The team used genetics and biochemistry to shed light on the role of the two proteins that they named SegA and SegB for chromosome segregation. “It was relatively straightforward to characterize SegA, as the protein contains a distinctive signature, known as Walker motif,” Barillà explains. “This consists of a short stretch of building blocks or amino acids that bind a small ligand called ATP. We’ve shown that SegA is able to bind ATP and to convert it into a smaller molecule, ADP.” While the binding of ATP and the conversion into ADP are crucial activities for the function of SegA, understanding the role of SegB was trickier: they suspected that this protein might be a DNA-binding protein. However, the problem was to find the site that the protein potentially bound to on the chromosome.
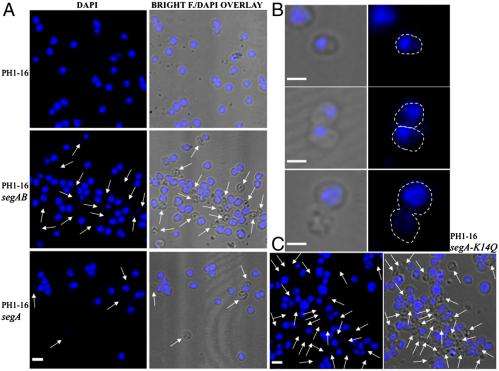
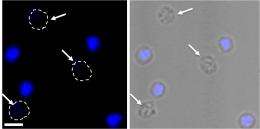
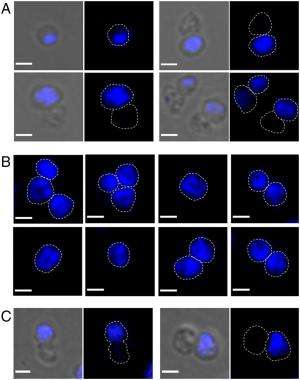
“We made an educated guess based on what is known for bacterial DNA segregation proteins and found out that SegB binds to specific sites on the chromosome,” she continues. “We employed microscopy to visualize cells in which we induced, if you will, an overdose of SegA and SegB, which resulted in numerous cells without chromosomes. When we induced a corresponding overdose of the DNA-binding protein SegB only, we observed fragmented, split chromosomes.” These findings indicated that the SegAB complex is involved in chromosome segregation in the thermophilic, or heat-loving, archaeon S. solfataricus.
One of the team’s key insights was the discovery that SegA polymerizes into filamentous structures upon binding ATP together with the finding that SegB promotes SegA assembly into polymers. “To investigate this aspect,” Barillà recounts, “we used dynamic light scattering. This method allowed us to determine the size of particles in solution on the basis of the amount of light that the particles scatter.” In fact, when a beam of laser light hits molecules in solution, each molecule will scatter back a certain amount of light that is proportional to its size: the larger the molecule, the higher the intensity of scattered light. “This technique is great, as it allowed us to detect SegA polymerization in real time. As soon as the protein binds the small ligand ATP, it instantaneously grows into polymers – that is, long particles scattering a large amount of light. It’s quite exciting to see this process in real time, while it’s actually happening.”
Going forward, Barillà points out, there’s a lot that can be done to investigate the nature and dynamics of the SegA polymers with dynamic light scattering. “In parallel, we’d like to visualize the filaments using electron microscopy, which is able to provide high-resolution images of individual particles. Moreover, we’d like to further investigate the interplay between SegA and the partner SegB to understand how the latter protein affects SegA behavior. The fact that SegA assembles into filaments in vitro suggests that in vivo it may form cytoskeletal structures involved in moving and delivering newly duplicated chromosomes to specific subcellular locations – so that, when the cell divides, each daughter cell inherits one chromosome. Therefore, we also intend to examine the localization of the proteins in S. solfataricus cells to shed light on what happens in vivo.”
Barillà also points out that in silico modeling is a possible avenue of investigation. “However,” she adds, “I think that this step would be a bit premature at this stage, as we need to learn more about this system. If we discover that SegA functions as a cytoskeletal motor protein in the cell, then a bioinformatic model would help to rationalize the potential dynamics of this factor within the cell.”
In addition, Barillà notes that the team’s discoveries are basic science findings that will not lead directly to new therapies for combating pathological conditions, because S. solfataricus and all the other members of the archaea phylum are non-pathogenic microorganisms that cause no infectious diseases. However, she adds, from an evolutionary and biotechnological standpoint, archaea are a terrific and exciting group of organisms. “They’ve generated considerable interest because of their ability to adapt to life under extreme conditions like very high or low temperatures, very acidic and alkaline pH and high salinity. Their unusual properties make these organisms a valuable and, so far, under-exploited resource in the development of novel biotechnological processes.” Potential industrial applications span from thermostable proteolytic enzymes to food-processing enzymes, from biomining to cellulose degrading enzymes, from bioremediation to the use of archaeal liposomes as carrier vehicles in vaccine formulation, or as delivery systems for drugs or genes.
“Going back to our findings,” Barillà concludes, “I think that SegA is an interesting object from a biotechnology viewpoint, as it is extremely thermostable – it remains a nicely folded and functional protein at high temperatures – and is able to polymerize. These two properties make SegA an interesting biocompatible material for a new generation of scaffolds for tissue engineering.”
More information: Chromosome segregation in Archaea mediated by a hybrid DNA partition machine, Published online before print February 21, 2012, PNAS March 6, 2012 vol. 109 no. 10 3754-3759, doi: 10.1073/pnas.1113384109
Copyright 2012 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.

















