New Technology Expected To Help Rapidly Identify Avian Flu Strains
Scientists from the University of Colorado at Boulder and the Centers for Disease Control and Prevention in Atlanta have developed a novel "gene chip" based on a single influenza virus gene that is expected to allow scientists to quickly identify specific flu viruses, including avian influenza H5N1.
The team used the gene chip, known as the MChip, to detect H5N1 samples collected over a three-year period from people and animals from diverse locations around the world, said Professor Kathy Rowlen of CU-Boulder's chemistry and biochemistry department, lead author on the study. In tests on 24 H5N1 viral isolates--or samples grown in the laboratory--as well as additional patient samples that tested positive for the common influenza viruses H3N2 and H1N1, the MChip provided accurate information on flu types and subtyples about 97 percent of the time, according Rowlen.
A paper on the work, being published online this week, is scheduled to appear in the Dec. 15 issue of the American Chemical Society's journal, Analytical Chemistry. Co-authors include CU-Boulder Professor Robert Kuchta, CU-Boulder postdoctoral researchers Erica Dawson, Chad Moore, Daniela Dankbar, Martin Mehlman and Michael Townsend, CU-Boulder doctoral student James Smagala, and Catherine Smith and Nancy Cox from the CDC.
The research was funded by the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health.
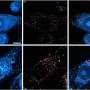
Based on the technology of the Flu Chip developed several years ago by a CU-Boulder team led by Rowlen and Kuchta, the MChip fits on a slide and contains an array of microscopic spots. Genetic bits of information complementary to H5N1 and other flu types and subtypes are placed robotically on the microarray, which is immersed in influenza gene fragments obtained from fluids of infected individuals.
RNA fragments from the infected fluid bind to specific DNA segments on the microarray like a key in a lock, indicating a match and that the virus signature is present. Since the RNA that binds to the microarray also contains a fluorescent dye, such "hits" light up when the chip is inserted into a laser scanner. An automated, artificial "neural network" trained to recognize the distinctive genetic pattern of H5N1 eliminates the possibility of human error, said Rowlen.
While the Flu Chip technology was based on three influenza genes, the MChip is based on a single gene segment that mutates less often and as a result may not need to be updated as frequently as flu genes that are typically used in diagnostic tests, Rowlen said. "The huge improvement here is that we need to amplify only one gene segment, making this a much easier and much more reliable process," she said.
Another potential advantage is that the MChip creates a way to simultaneously screen large numbers of flu samples to learn both the type and subtype of virus present, said Rowlen. While current real-time tests provide information about types of flu virus -- including type A and type B -- in a given sample, additional, time-consuming tests must now be run to determine subtypes like H5N1, she said.
The research team extracted H5N1 genetic materials from virus samples derived from human, feline and several types of avian hosts including geese, chicken and ducks. The samples, which were tested in biologically secure labs at the CDC in Atlanta, represented infections that had occurred between 2003 and 2006 over vast geographic areas, including Vietnam, Nigeria, Indonesia and Kazakhstan.
Virus diversity in such samples is critical, said Kuchta, because any diagnostic tool created for use on a rapidly changing virus like H5N1 must be able to detect as many variants as possible.
The team tested the ability of the MChip to correctly identify 24 different H5N1 viral isolates and distinguish those from seven non-H5N1 viral isolates, according to the study. The MChip accurately identified and gave complete subtype information for 21 out of the 24 strains of H5N1. The test gave no false positives, meaning the chip never indicated the presence of H5N1 when none was present.
"Our tests show the MChip can determine the type and subtype of human flu influenza in less than seven hours, in contrast to current methods to identify the type and subtype of flu that require several days," said Kuchta. "A new and rapid analysis like this should be particularly important for helping to limit the spread of new pandemic influenza strains."
Discussions are under way to commercialize the MChip, which costs less than $10 to manufacture from raw materials, said Rowlen. "One of our goals has been to address the needs of developing nations by providing an inexpensive field-portable test kit for the World Health Organization for global screening of respiratory illnesses."
While influenza A virus kills about 35,000 Americans every year, far less than 1 percent of those infected with it die, said Kuchta. In contrast, the avian influenza virus appears to kill about 50 percent of the people it infects. Currently, avian flu primarily infects and kills birds, but occasionally can be transferred to humans.
Source: University of Colorado at Boulder