March 2, 2007 feature
For low-cost DNA nanostructures, recycle sticky ends
Scientists from Duke University have recently demonstrated a new method for assembling large, low-cost DNA nanostructures, in part by reusing the “sticky-ends,” the broken DNA strands used to connect the nanostructures. In their hierarchical self-assembly method, the scientists have demonstrated one of the largest programmable synthetic nanostructures ever synthesized.
In their recent paper in Nanotechnology, Constantin Pistol and Chris Dwyer explain DNA’s advantages over silicon in terms of its easy synthesis, control and asymmetrical complexity. While recent advances in the field of fabricating DNA nanostructures have produced rigid sequences, Pistol and Dwyer focused on developing a self-assembly method that enables simple and inexpensive scaling of the DNA structures. In addition, their grid-like structures consist of components that can be independently modified to create arbitrary patterns for different purposes.
“The independent chemical modification of the motifs in the grid is important when generating templates on the grid,” Dwyer told PhysOrg.com. “Our computer architecture research is investigating how the DNA grids might act like scaffolds for other materials, such as carbon nanotubes. Arbitrary patterns of these materials are necessary to implement the kinds of circuits that we use to build computers.”
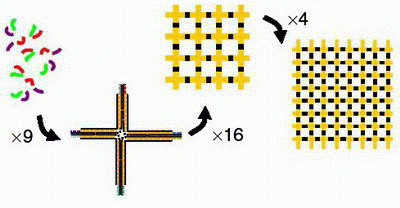
In their study, the scientists use a hierarchical approach: nine oligonucleotides (the structural units of DNA) form a cross-like motif, and 16 motifs annealed with streptavidin can form a grid. To build larger structures from these 4 x 4 grids, the scientists experimented with using two types of sticky-ends.
“Sticky-ends are single-stranded pieces of DNA that can hybridize, or form a double helix, with their complements,” Dwyer explained. “A well designed sticky-end has only one true complementary sticky-end in the system.”
The first type, the “generic” sticky-end, binds with only one helix instead of the normal two. This binding provides a relatively unstable interaction, which makes it easier to individually program two adjacent grids later on. The second type, the “specific” sticky-end, provides a stronger interaction and can control the weak interactions between the generic sticky-ends.
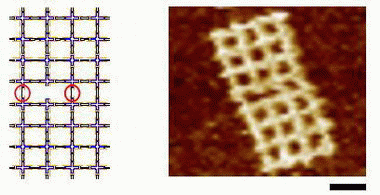
In one fabrication method, the scientists bound together two 4 x 4 grids, using two generic sticky-ends and one specific sticky-end (the fourth grid arm was left open for identification purposes). The scientists found that the single specific sticky-end could dominate the entire connection, providing a scalable assembly method.
In the second method, Pistol and Dwyer used all specific sticky-ends to connect two 4 x 4 grids. They found that, after the grids were connected, the sticky-ends could be reused in other connections. In this method, the scientists assembled four 4 x 4 grids to produce a 64-motif structure.
“Once the two sticky-ends have joined into a double helix, their sequences can be re-used in another part of the system,” said Dwyer. “This requires that the nanostructure be formed hierarchically.”
At a molecular weight of 8960 kD, the 64-motif structure is one of the largest programmable synthetic nanostructures ever synthesized. The scientists also predict that this method can be scaled even further before reaching a limit imposed when the generic interactions begin to dominate the process. However, studies in periodic DNA crystal formation suggest that the scale limit is nearly macroscopic.
In analyzing their structures for defects, Pistol and Dwyer found that missing motifs were common, requiring defect-tolerant designs in future large-scale assemblies. If DNA nanostructures can be fabricated at the scales Pistol and Dwyer predict, then not only biotechnology and the pharmaceutical industry would benefit, but future computer systems may use DNA, as well.
“One of the benefits of DNA nanostructures for computers is device density,” Dwyer said. “The grid has a pitch of 20nm, and this is about half of the smallest device feature in Intel's latest lithography process. The other benefit is manufacturing scale. Each experiment created a vast number of structures (~1012 or more) and this holds the promise of more complex and higher performance computers in the future.”
Citation: Pistol, Constantin, and Dwyer, Chris. “Scalable, low-cost, hierarchical assembly of programmable DNA nanostructures.” Nanotechnology 18 (2007) 125305 (4pp).
Copyright 2007 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.